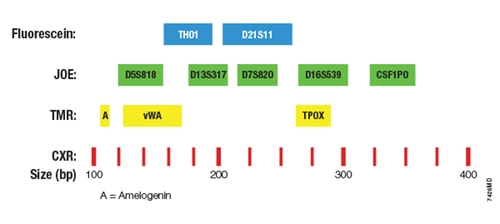
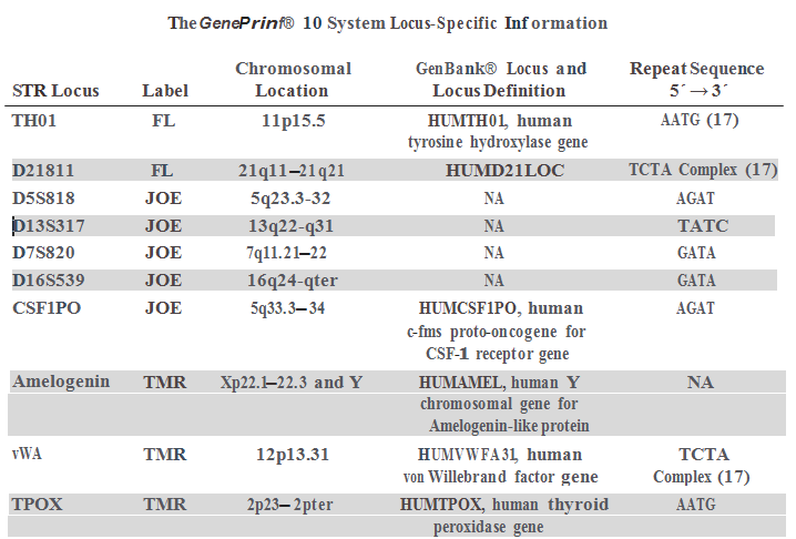
Genetic lociThe Promega GenePrint® 10 System coamplifies 10 human loci including 9 polymorphic tetranucleotide short tandem repeats (STR) and the Amelogenin gene for gender identification. The PCR amplicons, each less than 500 bp, are labeled with fluorescein (FL), 6-carboxy-4',5'-dichloro-2',7'-dimethoxyfluorescein ( JOE) or carboxy-tetramethylrhodamine (TMR).
Fig.1 The GenePrint® 10 System allows co-amplification and three-color detection of 10 loci, including all ASN-0002-2011 approved markers (TH01, TPOX, vWA, CSF1PO, D16S539, D7S820, D13S317, D5S818) plus Amelogenin and D21S11.
Adapted from Promega GenePrint ® 10 System Technical Manual TM392.
The loci selected by Promega for the assay have a high degree of heterozygosity and produce a minimum of artifacts associated with Taq DNA polymerase such as repeat slippage and terminal nucleotide addition. The primers selected for each locus generate amplified products within a limited size range allowing simultaneous detection of several loci without overlap of alleles across loci. DNA Amplification & Fragment AnalysisDNA submitted for analysis is requantified spectrophotometrically to verify sample concentration and purity. The assay is very sensitive and designed to perform optimally at a particular concentration range. Four nanograms of DNA from each sample are amplified by PCR in duplicate to verify the reproducibility of the results. All nine STR loci and the Amelogenin gene are coamplified in a single multiplex reaction. With every batch of samples analyzed, an aliquot of the 2800M control DNA (included with the Promega kit) is included, amplified, and genotyped for quality control to assure that the assay is performing as expected. A no-template control is also included in every run to detect reagent contamination.
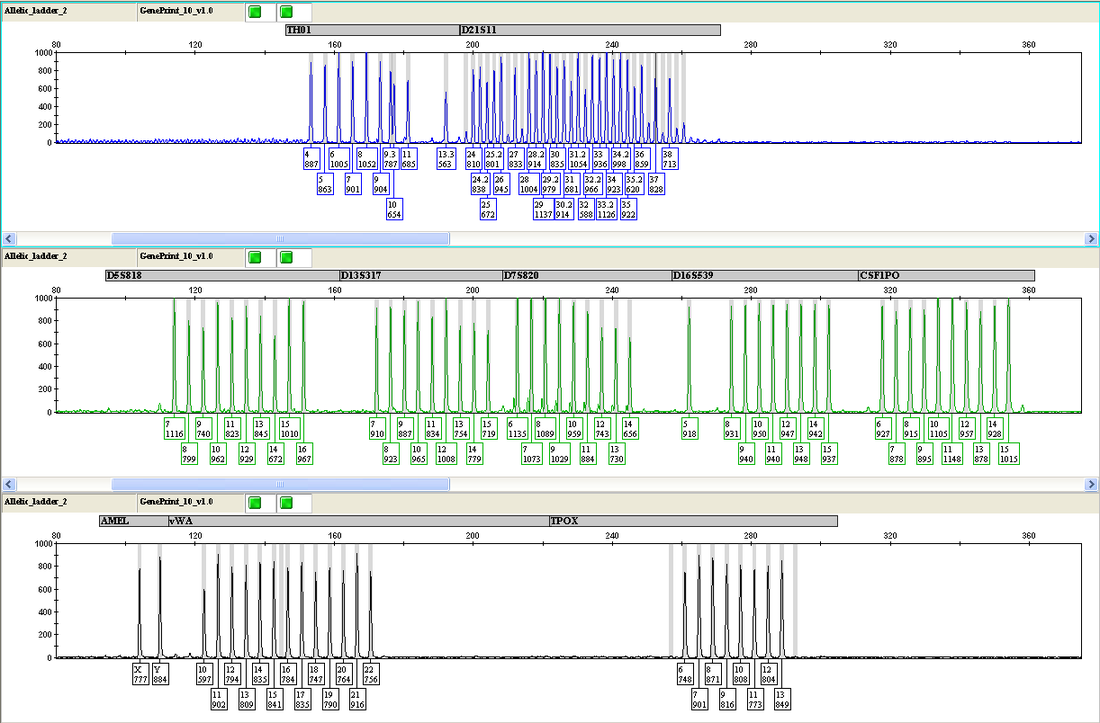
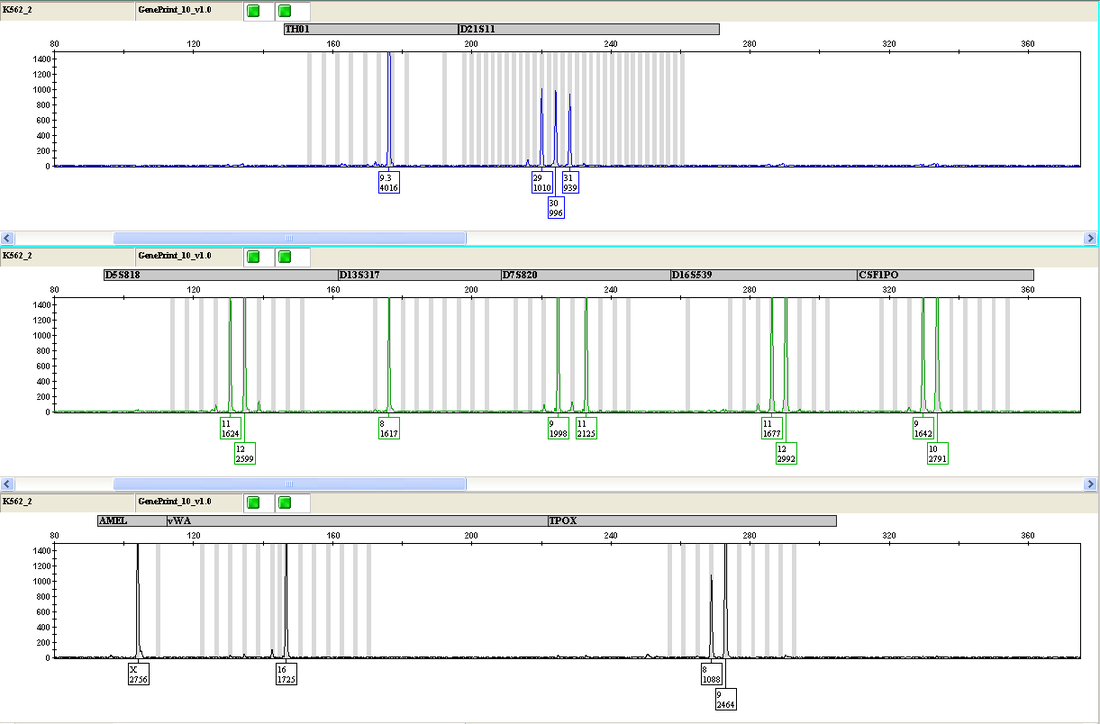
After PCR, each sample is loaded onto the Applied Biosystems 3130 xl Genetic Analyzer for separation, detection and analysis of the fluorescent amplified fragments. A fluorescent ladder (60-600 bases), labeled with carboxy-X-rhodamine, is placed in each sample and serves as an internal size standard to increase precision in the analyses. The resolved DNA fragments for each sample are compared to allelic ladders containing all known variants at each locus, which are included as a separate sample in every run (see Figure 1). The allelic ladders support the interpretation of data, and STR profiles are assigned to every sample with the assistance of Applied Biosystems GeneMapper® 4.0 software. Figure 1. GeneMapper output - Allelic ladder and the human chronic myelogenous leukemia cell line K562 The top 3 rows display the allelic ladder. The bottom 3 rows display the STR profile for cell line K562.
|
HOME | SERVICES | APPLICATION FOR SERVICES | CONTACT
©2021 Molecular Diagnostics Laboratory at Dana-Farber Cancer Institute
©2021 Molecular Diagnostics Laboratory at Dana-Farber Cancer Institute